Optimizing protein production in the One-Pot Pure system: insights into reaction composition and expression efficiency
Published in ACS Synthetic Biology, 2025
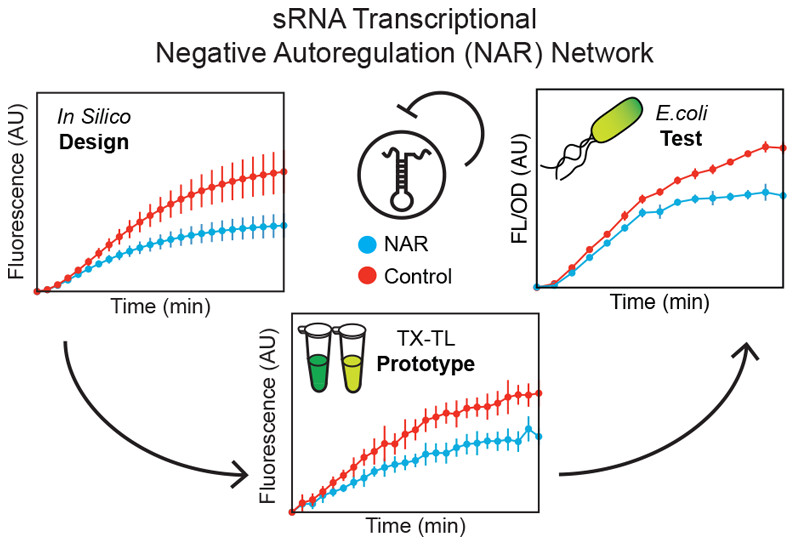
The One-Pot PURE (Protein synthesis Using Recombinant Elements) system simplifies the preparation of traditional PURE systems by coculturing and purifying 36 essential proteins for gene expression in a single step, enhancing accessibility and affordability for widespread laboratory adoption and customization. However, replicating this protocol to match the productivity of traditional PURE systems can take considerable time and effort due to uncharacterized variability. In this work, we observed unstable PURE protein expression in the original One-Pot PURE strains, E. coli M15/pREP4 and BL21(DE3), and addressed this issue using glucose-mediated catabolite repression to minimize burdensome background expression. We also identified several limitations making the M15/pREP4 strain unsuitable for PURE protein expression, including coculture incompatibility with BL21(DE3) and uncharacterized proteolytic activity. We showed that consolidating all expression vectors into a protease-deficient BL21(DE3) strain minimized proteolysis, led to more uniform coculture cell growth at the time of induction, and improved the stoichiometry of critical translation initiation factors in the final PURE mixture for efficient cell-free protein production. In addition to optimizing the One-Pot PURE protein composition, we found that variations in commercial energy solution formulations could compensate for suboptimal PURE protein stoichiometry. Notably, altering the source of E. coli tRNAs in the energy solution alone led to significant differences in the expression capacity of cell-free reactions, highlighting the importance of tRNA codon usage in influencing protein expression yield. Taken together, this work systematically investigates the proteome and biochemical factors influencing the One-Pot PURE system productivity, offering insights to enhance its robustness and adaptability across laboratories. 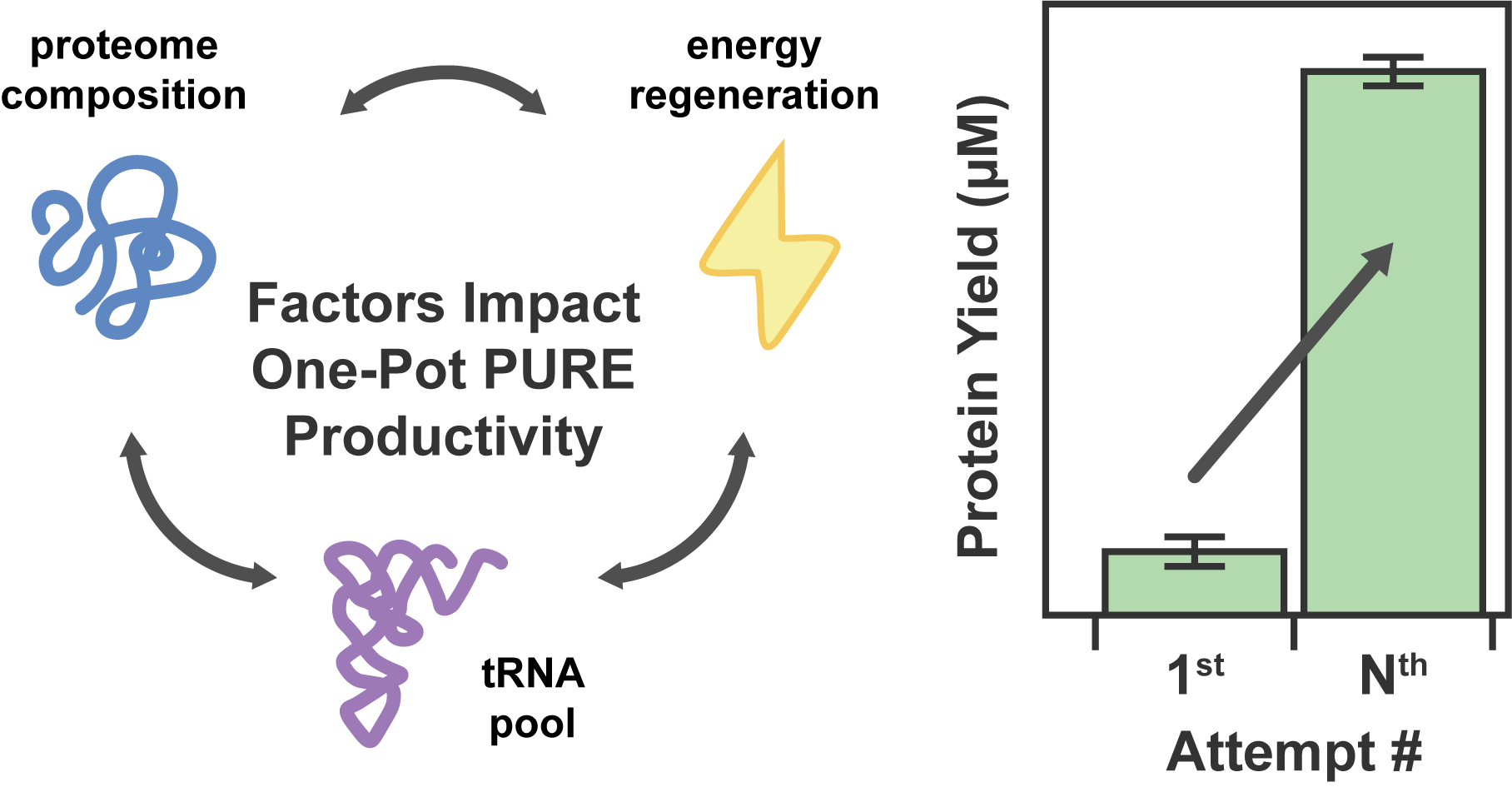
Recommended citation: Yan Zhang, Matas Deveikis, Yanping Qiu, Lovisa Björn, Zachary A. Martinez, Tsui-Fen Chou, Paul S. Freemont, Richard M. Murray, " Optimizing protein production in the One-Pot Pure system: insights into reaction composition and expression efficiency. " ACS Synthetic Biology, 2025.
Download Paper